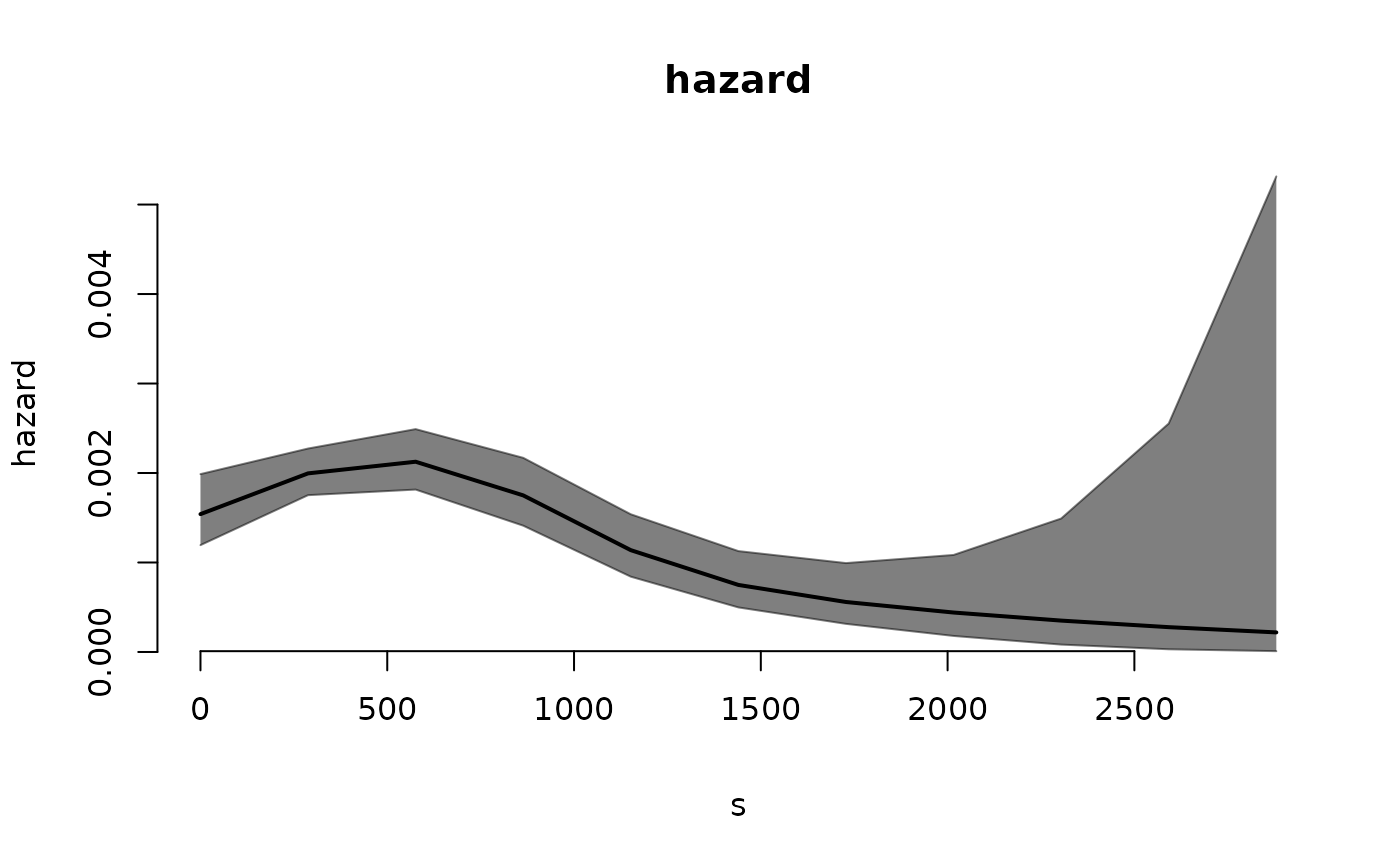
plot.haz1tsLMM() is a plot method for objects of class haz1tsLMM.
Arguments
- x
The output of the function
fit1ts.- which_plot
The type of plot required. Can be one of
"hazard"(default) or"covariates".- plot_grid
(optional) A named vector containing the parameters to build a new grid of intervals over
sfor plotting the estimated hazard on a finer grid. This must be of the form:plot_grid = c(smin, smax, ds), wheresmin,smaxare the minimum and maximum values desired for the intervals overs, anddsis the distance between intervals overs. If not specified, the plotting is done using the same B-splines basis as for the estimation. The function will check if the parameters for the grid provided by the user are compatible with those originally used to construct the B-splines for estimating the model. If not, the grid will be adjusted accordingly and a warning will be returned.- plot_options
A list with all possible options for any of the plots:
loghazardA Boolean. Default isFALSE. IfFALSEthe function returns a plot of the hazard curve, ifTRUEthe function returns a plot of the log-hazard curve.log10hazardA Boolean. Default isFALSE. IfTRUEit returns a plot of the log10-hazard curve.colThe color of the curve plotted. Default is"black".add_CIA Boolean. IfTRUE(default) the confidence bands will be added.col_CIThe color for the confidence bands. The default is the same color of the curve, with a 50% transparency level.mainThe title of the plot.xlabThe label of the time axis (plotted on the x axis).ylabThe label of the y-axis (hazard, log-hazard or log10-hazard).xlimA vector with two elements defining the limits of the time scale on the x axis.ylimA vector with two elements defining the limits of function plotted on the y axis (hazard, log-hazard or log10-hazard).xminThe minimum value on the x-axis.yminThe minimum value on the y-axis.cex_mainThe magnification to be used for the main title, default is 1.2 .cex_labThe magnification to be used for the axis labels, default is 1 .HRA Boolean. IfTRUEthe HRs with their CIs will be plotted. Default isFALSE(plot thebetawith their CIs).symmetric_CIA Boolean. Default isTRUE. If a plot of the HRs is required (HR == TRUE), then plot symmetrical Confidence Intervals, based on the SEs for the HRs calculated by delta method. IfFALSE, then CIs are obtained by exponentiating the CIs for the betas.confidenceThe level of confidence for the CIs. Default is .95 (alpha = 0.05).col_betaThe color for the plot of the covariates' effects.pchThe symbol for plotting the point estimates.
- ...
Further arguments to plot.
Details
The function obtainSmoothTrend from the R-package LMMsolver is
used here. We refer the interested readers to https://biometris.github.io/LMMsolver/
for more details on LMMsolver and its usage.
Examples
## preparing data - no covariates
dt1ts <- prepare_data(data = reccolon2ts,
s_in = "entrys",
s_out = "timesr",
events = "status",
ds = 180)
## fitting the model with fit1ts() - default options
mod1 <- fit1ts(dt1ts,
optim_method = "LMMsolver")
plot(mod1)